3rd Generation Sequencing Improves Fungal Barcoding
Improvements in DNA sequencing has impacted research in many ways.
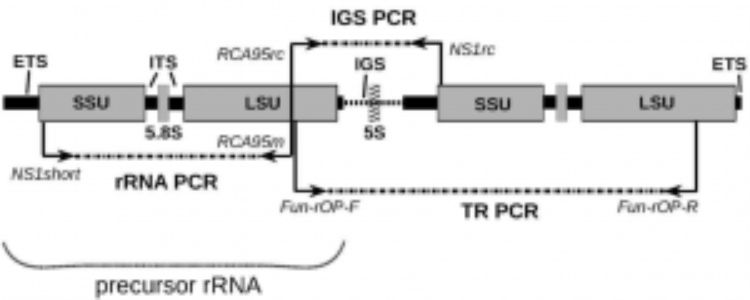
Improvements in DNA sequencing has impacted research in many ways. Known as “3rd generation” sequencing platforms, PacBio and Oxford Nanopore technologies allow sequencing of very long reads (>100kb) compared to classical Sanger Sequencing (<800bp). Common DNA regions that are sequenced for identifying fungi to a species level include the external transcribed spacer (ETS), small ribosomal subunit (SSU), internal transcribed spacer 1 (ITS1), 5.8S ribosomal RNA sequence (5.8S), internal transcribed spacer 2 (ITS2), large ribosomal subunit (LSU), and intergenic spacer (IGS). These sequences are very close together, and are often found clustered in tandem repeats in fungal genomes known as a ribosomal operon (Fig. 1).
A recent report in Molecular Ecology Resources by Wurzbacher et al. shows the power of 3rd generation sequencing technologies over Sanger Sequencing. By performing PCR and sequencing the entire ribosomal operon (~10kb) instead of sequencing each region individually through Sanger Sequencing, scientists can gain high quality data in a high throughput manner, providing clarity and resolution of fungal species relatedness. They test this new method on herbarium samples of Basidiomycetes, aquatic Chytridiomycetes, and cryptic single-celled fungi in the Nephridiophagidae family
The entire article can be found below:




 Print
Print Email
Email